The following list gives you the steps to use BTK in a Python environment.
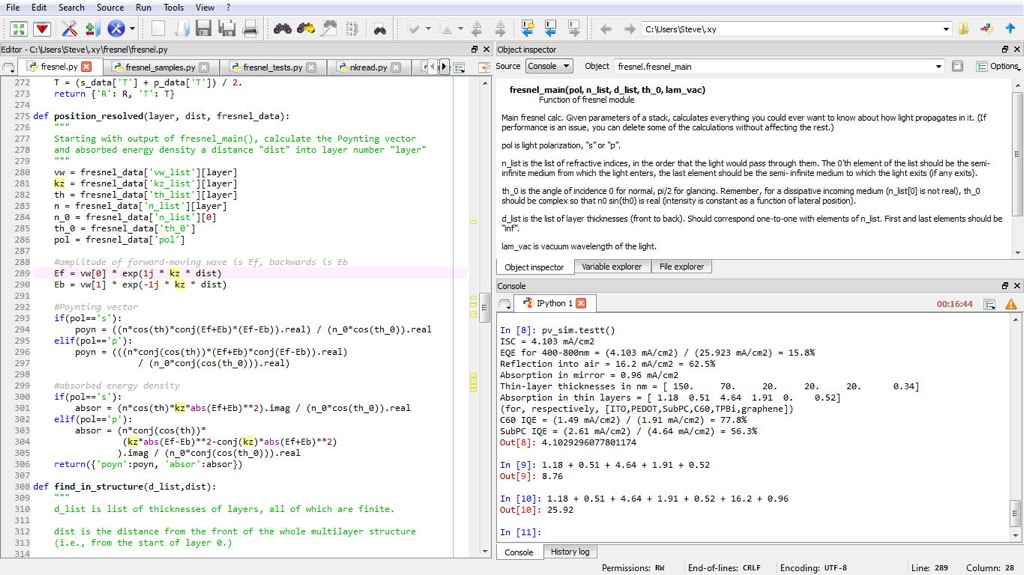
Python is a high-level language, free to use, running on different environments (Windows, Linux, MacOS X). It favours the object-oriented programming, which do not prevent to implement functional programming. The syntax is clear, doing python easy to learn and faster to develop. The popularity of Python increases in the scientific community. Indeed, lot of libraries can respond to any scientifical expectations (signal processing (Scipy) , 3D modelisation (pyVtk) , optimization (pyOpt), etc). Moreover, advanced interactive- development environment, like spyder (see figure below), appears allowing to easily switch from Matlab.
For further informations about Python versus matlab comparison, you can read this article.
If you put the two files (btk.py and btk.pyd (Windows), btk.so (Linux/MacOS X)) in the working directory, you only have to add the following code: import btk in you script header.
Otherwise, if the package is located in another folder then you need to add the path in the known path by Python.
The primary advantage of BTK is to offer lots of methods for handling a motion capture file, especially the C3D extension. This binary file embeds four containers:
Reading an acquisition is simple.
First of all, BTK prevents to extract point frame number and point-aquisition frequency from the metadata. Special accessors are proposed:
Now, let’s imagine that your acquisition only contains the points named into the acquisition:
Except that the index begins at 0, accessing to element of a numpy array is similar to the matlab syntax. For help about Numpy slicing and equivalent matlab functions, you can read the following user guide Numpy for Matlab Users.
Extracting analog object is similar to point extraction. Generally, analog measures are acquired at high-speed frequency. This parameter can be accessed with the method btkAcquisiton::GetAnalogFrequency() as the number of frame with the accessors btkAcquisition::GetAnalogFrameNumber(). Imagine that the c3d contains the muscular activities labeled: Emg1, Emg2, Emg3,... The following code will explain how to extract their values:
Writing data into an acquisition is as easy as reading data. The package proposed dedicated methods to do it directly on an aquisition object.
The following code firsly explains how to modify one point value. Secondly, a new point will be appended into the acquisition. In this case, developpers can use different signatures to build a new btkpoint object. In this example two signatures are illustrated. Others are detailed in the Doxygen help (see documentation of the class btkPoint).
The syntax is equivalent to the above point section.
All modifications are definitely recorded into a C3D file ONLY IF the following lines end your script. This code consists in building a btkAcquisitionFileWriter object and linking it to the modified acquisition. The method btkAcquisitionFileWriter::SetFilename allows you either to overwrite the existing file or to create a new one.
With Python language, one interest of BTK is to take advantage of scipy modules to implement biomechanical processes.
An unavoidable process is to filter the data. To this end, the module scipy.signal presents a large panel of filtering methods, with a syntax close to Matlab. For example, this code show how to filter an analog data with a Butterworth filter.
The second biomechanical process takes as example show how to find the nearest rotation matrix from the movement of the pelvis cluster, according Challis, 1995 (A procedure for determining rigid body transformation parameters, Journal of Biomechanics, 28(6), pp. 733-737). The code manipulates method associated with a numpy array object and used the linear algebra module of Scipy.
8 data_FrameRef = np.array([acq.GetPoint(‚�ôLASI‚Äô).GetValues()[0,:],
acq.GetPoint(’RASI’).GetValues()[0,:],
acq.GetPoint(’LPSI’).GetValues()[0,:],
acq.GetPoint(’RPSI’).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))Ä�LASI‚Äô).GetValues()[0,:],
acq.GetPoint(’RASI’).GetValues()[0,:],
acq.GetPoint(’LPSI’).GetValues()[0,:],
acq.GetPoint(’RPSI’).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))ô�ASI‚Äô).GetValues()[0,:],
acq.GetPoint(’RASI’).GetValues()[0,:],
acq.GetPoint(’LPSI’).GetValues()[0,:],
acq.GetPoint(’RPSI’).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))LASI‚�ô).GetValues()[0,:],
acq.GetPoint(’RASI’).GetValues()[0,:],
acq.GetPoint(’LPSI’).GetValues()[0,:],
acq.GetPoint(’RPSI’).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))�).GetValues()[0,:],
acq.GetPoint(’RASI’).GetValues()[0,:],
acq.GetPoint(’LPSI’).GetValues()[0,:],
acq.GetPoint(’RPSI’).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))ô�.GetValues()[0,:],
acq.GetPoint(’RASI’).GetValues()[0,:],
acq.GetPoint(’LPSI’).GetValues()[0,:],
acq.GetPoint(’RPSI’).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))).GetValues()[0,:],
9 acq.GetPoint(‚�ôRASI‚Äô).GetValues()[0,:],
acq.GetPoint(’LPSI’).GetValues()[0,:],
acq.GetPoint(’RPSI’).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))Ä�RASI‚Äô).GetValues()[0,:],
acq.GetPoint(’LPSI’).GetValues()[0,:],
acq.GetPoint(’RPSI’).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))ô�ASI‚Äô).GetValues()[0,:],
acq.GetPoint(’LPSI’).GetValues()[0,:],
acq.GetPoint(’RPSI’).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))RASI‚�ô).GetValues()[0,:],
acq.GetPoint(’LPSI’).GetValues()[0,:],
acq.GetPoint(’RPSI’).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))�).GetValues()[0,:],
acq.GetPoint(’LPSI’).GetValues()[0,:],
acq.GetPoint(’RPSI’).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))ô�.GetValues()[0,:],
acq.GetPoint(’LPSI’).GetValues()[0,:],
acq.GetPoint(’RPSI’).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))).GetValues()[0,:],
10 acq.GetPoint(‚�ôLPSI‚Äô).GetValues()[0,:],
acq.GetPoint(’RPSI’).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))Ä�LPSI‚Äô).GetValues()[0,:],
acq.GetPoint(’RPSI’).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))ô�PSI‚Äô).GetValues()[0,:],
acq.GetPoint(’RPSI’).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))LPSI‚�ô).GetValues()[0,:],
acq.GetPoint(’RPSI’).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))�).GetValues()[0,:],
acq.GetPoint(’RPSI’).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))ô�.GetValues()[0,:],
acq.GetPoint(’RPSI’).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))).GetValues()[0,:],
11 acq.GetPoint(‚�ôRPSI‚Äô).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))Ä�RPSI‚Äô).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))ô�PSI‚Äô).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))RPSI‚�ô).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))�).GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))ô�.GetValues()[0,:]])
Frame = 40
data_FrameChosen = np.array([acq.GetPoint(’LASI’).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))).GetValues()[0,:]])
13 data_FrameChosen = np.array([acq.GetPoint(‚�ôLASI‚Äô).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))Ä�LASI‚Äô).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))ô�ASI‚Äô).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))LASI‚�ô).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))�).GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))ô�.GetValues()[Frame,:],
acq.GetPoint(’RASI’).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))).GetValues()[Frame,:],
14 acq.GetPoint(‚�ôRASI‚Äô).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))Ä�RASI‚Äô).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))ô�ASI‚Äô).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))RASI‚�ô).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))�).GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))ô�.GetValues()[Frame,:],
acq.GetPoint(’LPSI’).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))).GetValues()[Frame,:],
15 acq.GetPoint(‚�ôLPSI‚Äô).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))Ä�LPSI‚Äô).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))ô�PSI‚Äô).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))LPSI‚�ô).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))�).GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))ô�.GetValues()[Frame,:],
acq.GetPoint(’RPSI’).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))).GetValues()[Frame,:],
16 acq.GetPoint(‚�ôRPSI‚Äô).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))Ä�RPSI‚Äô).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))ô�PSI‚Äô).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))RPSI‚�ô).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))�).GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))ô�.GetValues()[Frame,:]])
# Difference with mean values
A = data_FrameRef-np.mean(data_FrameRef,axis=0)
B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
# transposition of the data
A = A.transpose()
B = B.transpose()
# matrix multiplication
# (Note : with a numpy array, it’s the dot method, not the multiplication operator)
C = np.dot(B,A.transpose())
# singular decomposition, called the svd method of the scipy/linalg module
P,T,Q = scipy.linalg.svd(C)
# computation of the nearest rotation matrix R
mat = np.array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
R = np.dot(P,np.dot(mat,Q.transpose()))).GetValues()[Frame,:]])
18 A = data_FrameRef-np.mean(data_FrameRef,axis=0)
19 B = data_FrameChosen-np.mean(data_FrameChosen,axis=0)
25 C = np.dot(B,A.transpose())
27 P,T,Q = scipy.linalg.svd(C)
29 mat = np.array([[ 1., 0., 0.],
31 [ 0., 0., scipy.linalg.det(np.dot(P,Q.transpose()))]])
32 R = np.dot(P,np.dot(mat,Q.transpose()))
 1.8.5
1.8.5